usethis::use_data_raw(name = "individual")Project Data, Structure & Paths
Project Management
Project Aims & Objectives
Project Data
We’re in our new project so the first thing we need to do is get the data we’ll be working with. This is a common start to any project where you start with a few data files. These might be generated through your own data collection, given to you by others or published data products and you might need to clean, wrangle and combine them together to perform your analysis.
Q: Where should I save my raw data files?
conventions: Data management
-
Store raw data in
data-raw/: raw inputs to any pre-processing, read only.
- Keep any processing scripts in the same folder
- Whether and where you publish data depends on size and copyright considerations.
-
Store analytical data in
data/: any clean, processed data that is used as the input to the analysis.
- Should be published along side analysis.
Setting up a data-raw/ directory
We start by creating a data-raw directory in the root of our project. We can use usethis function usethis::use_data_raw(). This creates the data-raw directory and an .R script within where we can save code that turns raw data into analytical data in the data/ folder.
We can supply a name for the analytical dataset we’ll be creating in our script which automatically names the .R script for easy provenance tracking. In this case, we’ll be calling it individual.csv so let’s use "individual" for our name.
✔ Setting active project to '/cloud/project'
✔ Creating 'data-raw/'
✔ Adding '^data-raw$' to '.Rbuildignore'
✔ Writing 'data-raw/individual.R'
• Modify 'data-raw/individual.R'
• Finish the data preparation script in 'data-raw/individual.R'
• Use `usethis::use_data()` to add prepared data to packageThe data-raw/individual.R script created contains:
## code to prepare `individual` dataset goes here
usethis::use_data(individual, overwrite = TRUE)We will use this file to perform the necessary preprocessing on our raw data.
However, in the mean time we will also be experimenting with code and copying code over to our individual.R script when we are happy with it. so let’s create a new R script to work in.
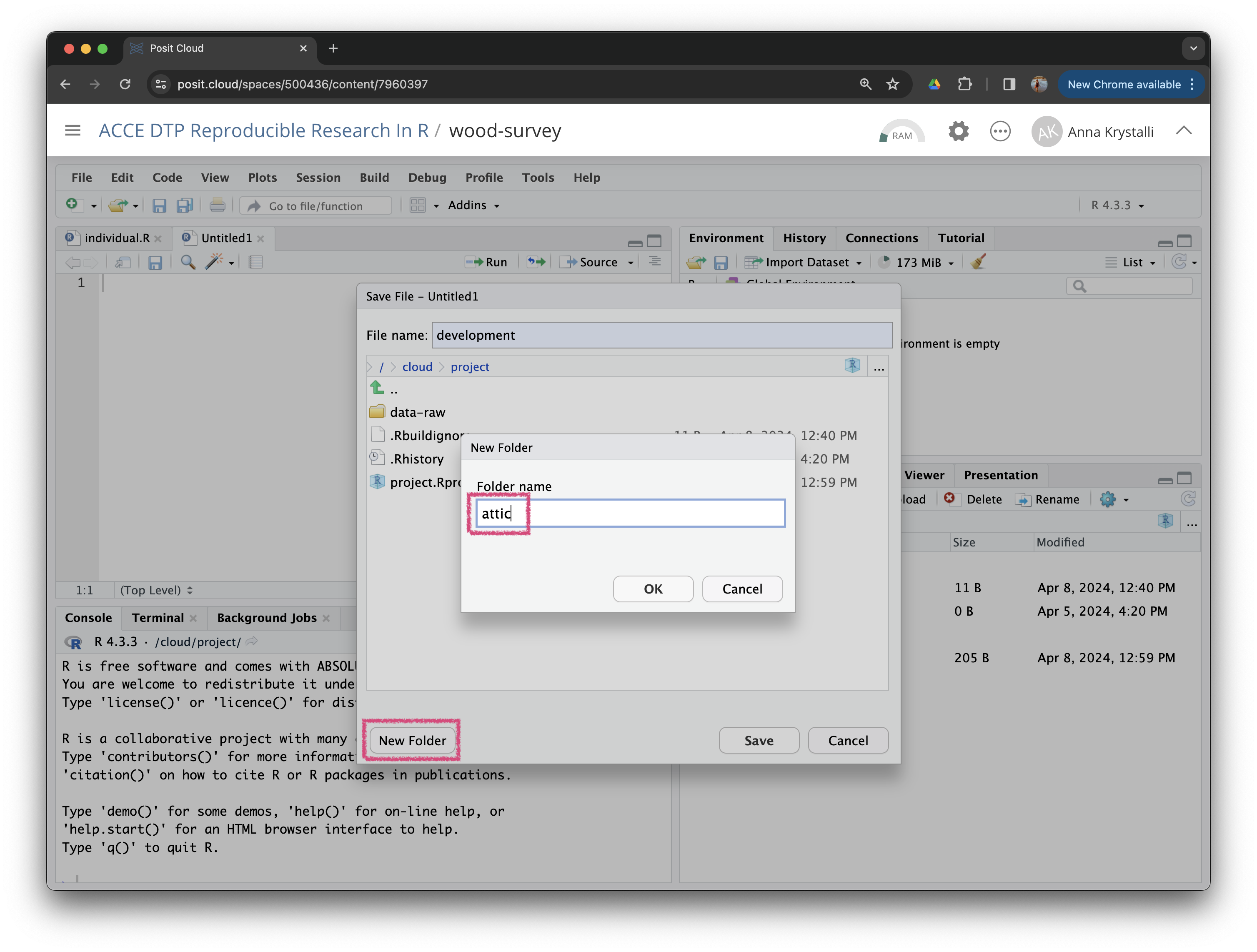
File > New File > R script
Let’s save this file in a new folder called attic/ and save it as file development.R.
Let’s work in development.R for now.
Download data
Now that we’ve got our data-raw folder, let’s download our data into it using function usethis::use_course() and supplying it with the url to the materials repository (bit.ly/wood-survey-data) and the path to the directory we want the materials saved into ("data-raw").
usethis::use_course("bit.ly/wood-survey-data",
destdir = "data-raw")✔ Downloading from 'https://bit.ly/wood-survey-data'
Downloaded: 7.61 MB
✔ Download stored in 'data-raw/wood-survey-data-master.zip'
✔ Unpacking ZIP file into 'wood-survey-data-master/' (77 files extracted)
Shall we delete the ZIP file ('wood-survey-data-master.zip')?
1: Nope
2: No way
3: I agree
Selection: 3
✔ Deleting 'wood-survey-data-master.zip'NEON Data
The downloaded folder contains a subset of data from the NEON Woody plant vegetation survey.
Citation: National Ecological Observatory Network. 2020. Data Products: DP1.10098.001. Provisional data downloaded from http://data.neonscience.org on 2020-01-15. Battelle, Boulder, CO, USA
This data product was downloaded from the NEON data portal and contains quality-controlled data from in-situ measurements of live and standing dead woody individuals and shrub groups, from all terrestrial NEON sites with qualifying woody vegetation.
Surveys of each site are completed once every 3 years.
Let’s have a look at what we’ve downloaded:
.
├── R
├── data-raw
│ ├── individual.R
│ └── wood-survey-data-master
│ ├── NEON_vst_variables.csv
│ ├── README.md
│ ├── individual [67 entries exceeds filelimit, not opening dir]
│ ├── methods
│ │ ├── NEON.DOC.000914vB.pdf
│ │ ├── NEON.DOC.000987vH.pdf
│ │ └── NEON_vegStructure_userGuide_vA.pdf
│ ├── vst_mappingandtagging.csv
│ └── vst_perplotperyear.csv
└── wood-survey.RprojThe important files for the analysis we want to perform are
├── individual [67 entries exceeds filelimit, not opening dir]
├── vst_mappingandtagging.csv
└── vst_perplotperyear.csv
vst_perplotperyear: Plot level metadata, including plot geolocation.
- one record per
plotIDpereventID, - describe the presence/absence of woody growth forms
- sampling area utilized for each growth form.
| uid | plotID | plotType | nlcdClass | decimalLatitude | decimalLongitude | geodeticDatum | easting | northing | utmZone | elevation | elevationUncertainty | eventID |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 93ee1436-cdd8-40bd-96c4-0585f36b904f | BART_002 | distributed | deciduousForest | 44.03508 | -71.27285 | WGS84 | 317882.0 | 4878281 | 19N | 550.8 | 0.4 | vst_BART_2016 |
| 4b5f972f-d00f-4766-b7d7-ae488e058416 | BART_003 | distributed | deciduousForest | 44.05525 | -71.26315 | WGS84 | 318720.5 | 4880500 | 19N | 439.5 | 0.3 | vst_BART_2016 |
| 66594b70-4db4-4005-bfc8-e42a1bdba15d | BART_006 | distributed | deciduousForest | 44.06051 | -71.31091 | WGS84 | 314911.2 | 4881190 | 19N | 432.7 | 0.2 | vst_BART_2016 |
| 730098e8-30a7-4b7a-a5ee-fde5318cc416 | BART_007 | distributed | mixedForest | 44.04970 | -71.29849 | WGS84 | 315873.0 | 4879961 | 19N | 388.4 | 0.2 | vst_BART_2016 |
| 07c96abe-6d78-4818-8b2e-f33fac4d06b5 | BART_010 | distributed | deciduousForest | 44.05007 | -71.26668 | WGS84 | 318422.2 | 4879932 | 19N | 430.2 | 0.2 | vst_BART_2016 |
| 557410ec-351d-4348-97e5-6dc0625c4f03 | BART_011 | distributed | mixedForest | 44.05001 | -71.29627 | WGS84 | 316051.2 | 4879991 | 19N | 370.1 | 0.2 | vst_BART_2016 |
vst_mappingandtagging: Mapping, identifying and tagging of individual stems for re-measurement.
- one record per
individualID. - data invariant through time, including
tagID,taxonIDand mapped location. - Records can be linked to
vst_perplotperyearvia theplotIDandeventIDfields.
| uid | eventID | pointID | stemDistance | stemAzimuth | individualID | taxonID | scientificName | taxonRank |
|---|---|---|---|---|---|---|---|---|
| 3a4301d5-8ff1-491f-bba7-e0a595ece6af | vst_BART_2015 | 43 | 13.1 | 341.7 | NEON.PLA.D01.BART.00101 | ACRU | Acer rubrum L. | species |
| 229a8489-dfef-4a50-9c2b-9bb4d614173e | vst_BART_2015 | 61 | 1.2 | 206.2 | NEON.PLA.D01.BART.00102 | ACRU | Acer rubrum L. | species |
| 27712596-d6d2-44e4-a462-cbdedef8a408 | vst_BART_2015 | 61 | 4.6 | 288.9 | NEON.PLA.D01.BART.00103 | FAGR | Fagus grandifolia Ehrh. | species |
| de648865-7d18-4a48-96ec-99265dc653ad | vst_BART_2015 | 57 | 30.3 | 94.7 | NEON.PLA.D01.BART.00106 | FAGR | Fagus grandifolia Ehrh. | species |
| 04c88265-7e34-4fd7-89ec-dc30a513c265 | vst_BART_2015 | 57 | 30.6 | 92.8 | NEON.PLA.D01.BART.00107 | FAGR | Fagus grandifolia Ehrh. | species |
| ff9975c3-c068-4d48-a27b-5175783d91f6 | vst_BART_2015 | 43 | 2.2 | 92.3 | NEON.PLA.D01.BART.00108 | FAGR | Fagus grandifolia Ehrh. | species |
vst_apparentindividual: Biomass and productivity measurements of apparent individuals.
- includes biomass, productivity and other measurements.
- may contain multiple records per individuals but only one record per
individualIDpereventID. - includes growth form, structure
- currently in separate files contained in
individual/ - may be linked to:
-
vst_mappingandtaggingrecords viaindividualID -
vst_perplotperyearvia theplotIDandeventIDfields.
-
| uid | namedLocation | date | eventID | domainID | siteID | plotID | individualID | growthForm | stemDiameter | measurementHeight | height |
|---|---|---|---|---|---|---|---|---|---|---|---|
| a36a162d-ed1f-4f80-ae45-88e973855c68 | BART_037.basePlot.vst | 2015-08-26 | vst_BART_2015 | D01 | BART | BART_037 | NEON.PLA.D01.BART.05285 | single bole tree | 17.1 | 130 | 15.2 |
| 68dc7adf-48e2-4f7a-9272-9a468fde6d55 | BART_037.basePlot.vst | 2015-08-26 | vst_BART_2015 | D01 | BART | BART_037 | NEON.PLA.D01.BART.05279 | single bole tree | 13.7 | 130 | 9.8 |
| a8951ab9-4462-48dd-ab9e-7b89e24f2e03 | BART_044.basePlot.vst | 2015-08-26 | vst_BART_2015 | D01 | BART | BART_044 | NEON.PLA.D01.BART.05419 | single bole tree | 12.3 | 130 | 7.7 |
| eb348eaf-3969-46a4-ac3b-523c3548efeb | BART_044.basePlot.vst | 2015-08-26 | vst_BART_2015 | D01 | BART | BART_044 | NEON.PLA.D01.BART.05092 | single bole tree | 12.1 | 130 | 15.2 |
| 2a4478ef-5970-40b6-b696-d1167cbe42ac | BART_044.basePlot.vst | 2015-08-26 | vst_BART_2015 | D01 | BART | BART_044 | NEON.PLA.D01.BART.05443 | single bole tree | 29.2 | 130 | 16.7 |
| e485203e-879e-4b56-b13a-0a6a56f0040f | BART_044.basePlot.vst | 2015-08-26 | vst_BART_2015 | D01 | BART | BART_044 | NEON.PLA.D01.BART.05432 | single bole tree | 12.1 | 130 | 10.6 |
As our first challenge, we are going to combined all the files in individual/ into a single analytical data file!
Paths
First let’s investigate our data. We want to access the files so we need to give R paths in order to load the data. We can work with the file system programmatically through R.
Creating portable paths with here
We’ll use the here package and function here() to create paths relative to the project root directory.
This is a good practice as it makes our code portable and independent of the where code is evaluated or saved.
What you never want to do is hard code paths in your code. This makes your code non-portable and can lead to errors when sharing code or moving code to a different machine or to a different location within a project.
Let’s start by creating a path to the downloaded data directory using here.
To create relative paths to files or directories with here() we provide character strings separated by commas that represent the path to the file or directory.
raw_data_path <- here::here("data-raw", "wood-survey-data-master")raw_data_path[1] "/cloud/project/data-raw/wood-survey-data-master"We can use raw_data_path as our basis for specifying paths to files within it. There’s a number of ways we can do this in R but I wanted to introduce you to package fs. It has a nice interface and extensive functionality for working with your file system programmatically.
fs::path(raw_data_path, "individual")/cloud/project/data-raw/wood-survey-data-master/individualLet’s now use function dir_ls to get a character vector of paths to all the individual files in directory individual.
/cloud/project/data-raw/wood-survey-data-master/individual/NEON.D01.BART.DP1.10098.001.vst_apparentindividual.2015-08.basic.20190806T172340Z.csv
/cloud/project/data-raw/wood-survey-data-master/individual/NEON.D01.BART.DP1.10098.001.vst_apparentindividual.2015-09.basic.20190806T144119Z.csv
/cloud/project/data-raw/wood-survey-data-master/individual/NEON.D01.BART.DP1.10098.001.vst_apparentindividual.2016-08.basic.20190806T143255Z.csv
/cloud/project/data-raw/wood-survey-data-master/individual/NEON.D01.BART.DP1.10098.001.vst_apparentindividual.2016-09.basic.20190806T143433Z.csv
/cloud/project/data-raw/wood-survey-data-master/individual/NEON.D01.BART.DP1.10098.001.vst_apparentindividual.2016-10.basic.20190806T144133Z.csv
/cloud/project/data-raw/wood-survey-data-master/individual/NEON.D01.BART.DP1.10098.001.vst_apparentindividual.2017-07.basic.20190806T144111Z.csvWe can check how many files we’ve got:
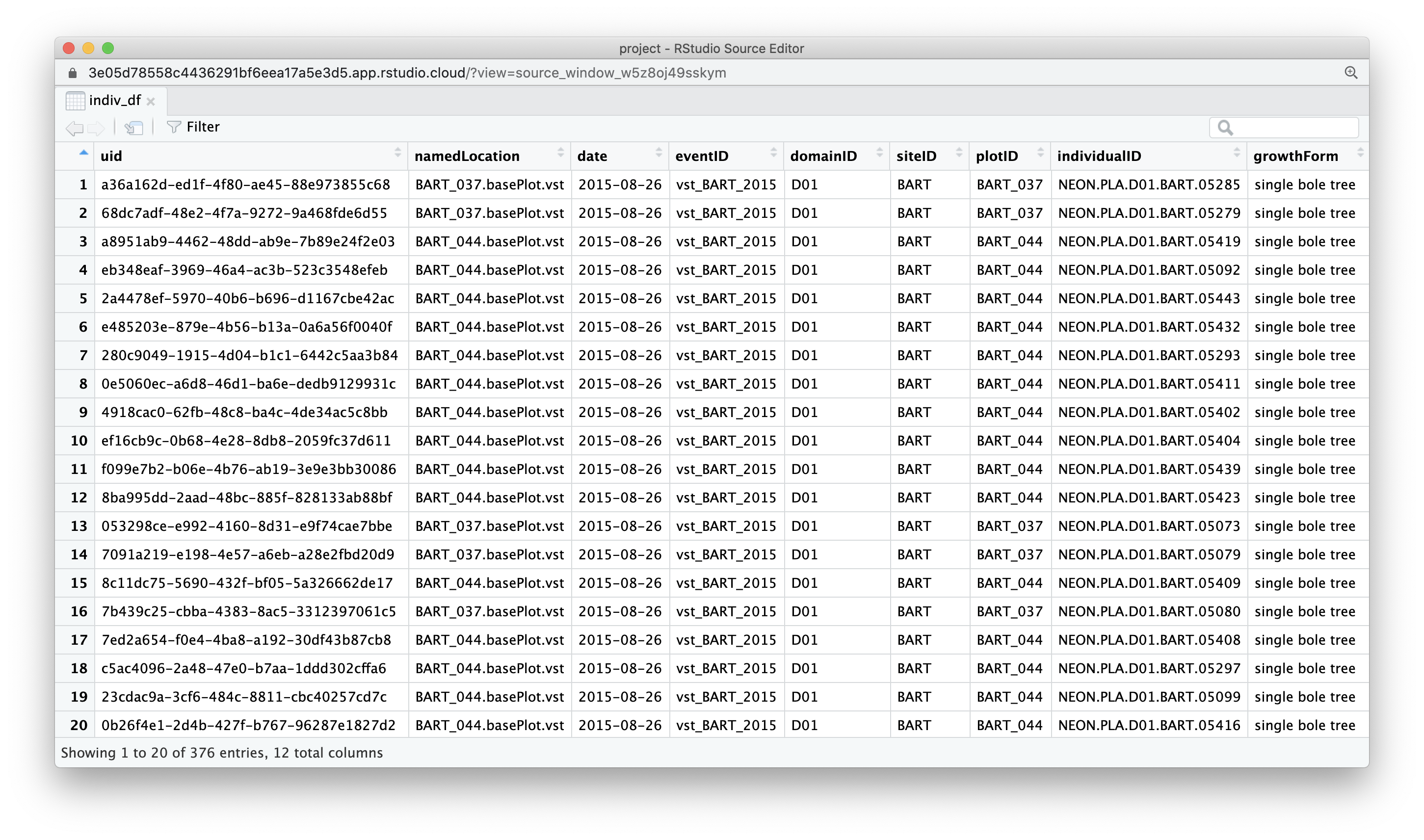
length(individual_paths)[1] 67We can now** use this vector of paths to read in files. Let’s read the first file in and check it out.** We use function read_csv() from readr package which reads comma delimited files into tibbles.
indiv_df <- readr::read_csv(individual_paths[1])Rows: 376 Columns: 12
── Column specification ────────────────────────────────────────────────────────
Delimiter: ","
chr (8): uid, namedLocation, eventID, domainID, siteID, plotID, individualI...
dbl (3): stemDiameter, measurementHeight, height
date (1): date
ℹ Use `spec()` to retrieve the full column specification for this data.
ℹ Specify the column types or set `show_col_types = FALSE` to quiet this message.indiv_df# A tibble: 376 × 12
uid namedLocation date eventID domainID siteID plotID individualID
<chr> <chr> <date> <chr> <chr> <chr> <chr> <chr>
1 a36a162… BART_037.bas… 2015-08-26 vst_BA… D01 BART BART_… NEON.PLA.D0…
2 68dc7ad… BART_037.bas… 2015-08-26 vst_BA… D01 BART BART_… NEON.PLA.D0…
3 a8951ab… BART_044.bas… 2015-08-26 vst_BA… D01 BART BART_… NEON.PLA.D0…
4 eb348ea… BART_044.bas… 2015-08-26 vst_BA… D01 BART BART_… NEON.PLA.D0…
5 2a4478e… BART_044.bas… 2015-08-26 vst_BA… D01 BART BART_… NEON.PLA.D0…
6 e485203… BART_044.bas… 2015-08-26 vst_BA… D01 BART BART_… NEON.PLA.D0…
7 280c904… BART_044.bas… 2015-08-26 vst_BA… D01 BART BART_… NEON.PLA.D0…
8 0e5060e… BART_044.bas… 2015-08-26 vst_BA… D01 BART BART_… NEON.PLA.D0…
9 4918cac… BART_044.bas… 2015-08-26 vst_BA… D01 BART BART_… NEON.PLA.D0…
10 ef16cb9… BART_044.bas… 2015-08-26 vst_BA… D01 BART BART_… NEON.PLA.D0…
# ℹ 366 more rows
# ℹ 4 more variables: growthForm <chr>, stemDiameter <dbl>,
# measurementHeight <dbl>, height <dbl>Run ?read_delim for more details on reading in tabular data.
Basic checks
Let’s perform some of the basic checks we learnt before we proceed.
View(indiv_df)names(indiv_df) [1] "uid" "namedLocation" "date"
[4] "eventID" "domainID" "siteID"
[7] "plotID" "individualID" "growthForm"
[10] "stemDiameter" "measurementHeight" "height" str(indiv_df)spc_tbl_ [376 × 12] (S3: spec_tbl_df/tbl_df/tbl/data.frame)
$ uid : chr [1:376] "a36a162d-ed1f-4f80-ae45-88e973855c68" "68dc7adf-48e2-4f7a-9272-9a468fde6d55" "a8951ab9-4462-48dd-ab9e-7b89e24f2e03" "eb348eaf-3969-46a4-ac3b-523c3548efeb" ...
$ namedLocation : chr [1:376] "BART_037.basePlot.vst" "BART_037.basePlot.vst" "BART_044.basePlot.vst" "BART_044.basePlot.vst" ...
$ date : Date[1:376], format: "2015-08-26" "2015-08-26" ...
$ eventID : chr [1:376] "vst_BART_2015" "vst_BART_2015" "vst_BART_2015" "vst_BART_2015" ...
$ domainID : chr [1:376] "D01" "D01" "D01" "D01" ...
$ siteID : chr [1:376] "BART" "BART" "BART" "BART" ...
$ plotID : chr [1:376] "BART_037" "BART_037" "BART_044" "BART_044" ...
$ individualID : chr [1:376] "NEON.PLA.D01.BART.05285" "NEON.PLA.D01.BART.05279" "NEON.PLA.D01.BART.05419" "NEON.PLA.D01.BART.05092" ...
$ growthForm : chr [1:376] "single bole tree" "single bole tree" "single bole tree" "single bole tree" ...
$ stemDiameter : num [1:376] 17.1 13.7 12.3 12.1 29.2 12.1 23.4 39.5 10 10.6 ...
$ measurementHeight: num [1:376] 130 130 130 130 130 130 130 130 130 130 ...
$ height : num [1:376] 15.2 9.8 7.7 15.2 16.7 10.6 18.4 19 5.7 8.7 ...
- attr(*, "spec")=
.. cols(
.. uid = col_character(),
.. namedLocation = col_character(),
.. date = col_date(format = ""),
.. eventID = col_character(),
.. domainID = col_character(),
.. siteID = col_character(),
.. plotID = col_character(),
.. individualID = col_character(),
.. growthForm = col_character(),
.. stemDiameter = col_double(),
.. measurementHeight = col_double(),
.. height = col_double()
.. )
- attr(*, "problems")=<externalptr> summary(indiv_df) uid namedLocation date eventID
Length:376 Length:376 Min. :2015-08-26 Length:376
Class :character Class :character 1st Qu.:2015-08-27 Class :character
Mode :character Mode :character Median :2015-08-27 Mode :character
Mean :2015-08-27
3rd Qu.:2015-08-31
Max. :2015-08-31
domainID siteID plotID individualID
Length:376 Length:376 Length:376 Length:376
Class :character Class :character Class :character Class :character
Mode :character Mode :character Mode :character Mode :character
growthForm stemDiameter measurementHeight height
Length:376 Min. : 2.00 Min. : 10.0 Min. : 0.50
Class :character 1st Qu.:13.90 1st Qu.:130.0 1st Qu.:10.60
Mode :character Median :20.20 Median :130.0 Median :14.30
Mean :23.01 Mean :129.5 Mean :13.91
3rd Qu.:29.55 3rd Qu.:130.0 3rd Qu.:17.23
Max. :69.90 Max. :130.0 Max. :30.20 skimr::skim(indiv_df)| Name | indiv_df |
| Number of rows | 376 |
| Number of columns | 12 |
| _______________________ | |
| Column type frequency: | |
| character | 8 |
| Date | 1 |
| numeric | 3 |
| ________________________ | |
| Group variables | None |
Variable type: character
| skim_variable | n_missing | complete_rate | min | max | empty | n_unique | whitespace |
|---|---|---|---|---|---|---|---|
| uid | 0 | 1.00 | 36 | 36 | 0 | 376 | 0 |
| namedLocation | 0 | 1.00 | 21 | 21 | 0 | 7 | 0 |
| eventID | 0 | 1.00 | 13 | 13 | 0 | 1 | 0 |
| domainID | 0 | 1.00 | 3 | 3 | 0 | 1 | 0 |
| siteID | 0 | 1.00 | 4 | 4 | 0 | 1 | 0 |
| plotID | 0 | 1.00 | 8 | 8 | 0 | 7 | 0 |
| individualID | 0 | 1.00 | 23 | 23 | 0 | 374 | 0 |
| growthForm | 3 | 0.99 | 7 | 16 | 0 | 4 | 0 |
Variable type: Date
| skim_variable | n_missing | complete_rate | min | max | median | n_unique |
|---|---|---|---|---|---|---|
| date | 0 | 1 | 2015-08-26 | 2015-08-31 | 2015-08-27 | 3 |
Variable type: numeric
| skim_variable | n_missing | complete_rate | mean | sd | p0 | p25 | p50 | p75 | p100 | hist |
|---|---|---|---|---|---|---|---|---|---|---|
| stemDiameter | 0 | 1 | 23.01 | 11.22 | 2.0 | 13.9 | 20.2 | 29.55 | 69.9 | ▆▇▃▁▁ |
| measurementHeight | 0 | 1 | 129.48 | 6.76 | 10.0 | 130.0 | 130.0 | 130.00 | 130.0 | ▁▁▁▁▇ |
| height | 0 | 1 | 13.91 | 4.45 | 0.5 | 10.6 | 14.3 | 17.22 | 30.2 | ▁▅▇▂▁ |
Update individual.R
Everything looks good. Before moving on, let’s update our individual.R script with the code we’ve just written and want to formally keep as part of out processing pipeline.
Add the following code and comments to the bottom of individual.R:
So let’s now move onto the next step of reading in all the files and combining them together. To do this, we’ll examine the principles of Iteration.